MARCH 11 2025
Building an intelligent clinical study search system
Discover how clinical research is transformed with AI-powered semantic search systems using Hypermode
In the rapidly evolving landscape of clinical research, finding relevant studies and statements across vast databases of medical documentation poses a significant challenge. This blog explores how healthcare technology companies can leverage Dgraph's graph engine and the Modus agentic API framework to build an intelligent semantic search system for clinical studies.
The challenge
Clinical research is the foundation of medical advancement, with thousands of studies generating vast amounts of critical data each year. However, finding relevant studies, comparing methodologies, and identifying patterns across trials remains a significant challenge. To help clients identify risk, reduce costs, and accelerate clinical development timelines, many life sciences companies want to modernize study design and help study sponsors deliver therapies to market faster.
Clinical studies often struggle to find participants and encounter delays, but knowledge graphs give them the ability to identify risks and opportunities, model operational trade-offs in real time and better see how study decisions will affect participants, sites and sponsors. Teams behind these clinical studies often need to manage a complex knowledge graph, where each study is connected to various qualifiers and ontological classifications. These connections represent important attributes like study phase, disease indications, interventions, and other clinical parameters.
The challenge that many encounter is to build a search system that can:
- Find similar clinical studies based on contextual information
- Match relevant statements across studies
- Handle structured and unstructured data
- Maintain data integrity with a large medical ontology
- Scale efficiently without duplicating data across multiple systems
Building the foundation for intelligent search
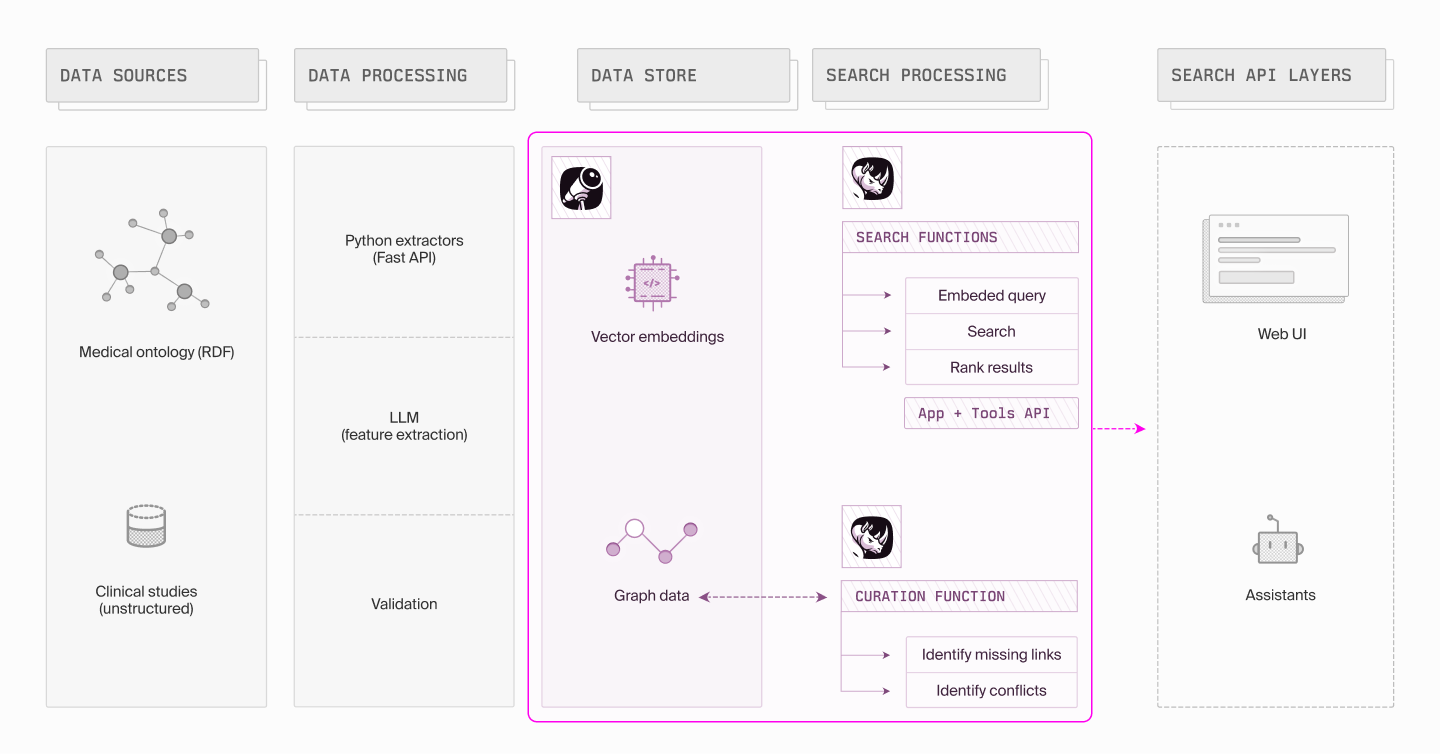
Building an intelligent search system for clinical studies requires carefully balancing multiple technical considerations. The system needs to handle both structured relationships between medical concepts and unstructured textual descriptions, while maintaining fast and accurate search capabilities. Let’s explore an approach that combines Dgraph's graph database capabilities with modern vector search techniques, integrated through the Modus framework to create a flexible and powerful search solution.
The data model
The system's foundation is built on Dgraph, given its ability to handle complex relationships in a graph structure that includes:
- Documents (clinical studies) as primary nodes
- Qualifier nodes that connect to ontology classes
- Key-value pairs representing study attributes (for example, study phase, indication)
- A large medical ontology with tens of thousands of terms
At the core of the data model are clinical study documents, which serve as primary nodes in the graph. Each study connects to multiple qualifier nodes, which in turn link to specific classes within a comprehensive medical ontology. This structure allows for precise representation of study attributes like phase, indication, and intervention type.
Medical ontologies tend to be extensive, containing tens of thousands of terms that cover diseases, treatments, methodologies, and other clinical concepts. Rather than forcing this complex, connected data into a table data structure using a traditional relational database system, which are difficult and costly to maintain, the system treats the ontology as a separate layer that integrates with the main graph structure. This approach provides flexibility while maintaining the semantic relationships crucial for accurate search results.
Implementing vector search
Dgraph's vector search capabilities help teams implement semantic similarity
search directly within the database. This is achieved through the use of the new
vfloat property type, which efficiently stores vector embeddings alongside the
structured data. These embeddings capture the semantic meaning of study
descriptions, titles, and other textual content.
To ensure fast similarity searches across millions of vectors, the system
employs Hierarchical Navigable Small World (HNSW) indexing. This advanced
indexing technique significantly reduces the computational cost of finding
similar vectors while maintaining high accuracy. The similar_to
DQL function provides a simple interface for
querying these vector embeddings, allowing for seamless integration of semantic
search with traditional graph traversal queries.
Ensuring rich context through the search pipeline
The search process is implemented as a sophisticated pipeline that combines multiple techniques to deliver relevant results. The pipeline begins with context collection, where the system gathers all relevant attributes of a study, including its phase, title, purpose, and associated qualifiers from the knowledge graph. This creates a rich context that goes beyond simple text matching.
The next step is query generation. Using Modus, the system converts structured study attributes into natural language representations that capture the semantic intent of the search. Large Language Models (LLM) play a key role here, generating sophisticated search queries that understand medical terminology and context and creating an embedding of the natural language description. This brings critical context that bridges structured data and natural language understanding.
The vector search phase compares these embeddings to the vectors stored in
Dgraph. Using the similar_to function, the system finds semantically similar
studies based on their distance in vector space. Importantly, this search can be
filtered based on structured attributes like study phase or indication,
combining the benefits of semantic and traditional search approaches.
Finally, the result processing stage applies sophisticated ranking algorithms that consider multiple factors. Results are re-ranked based on a combination of vector similarity scores, graph relationship strength, and specific business rules. This ensures that the most relevant results appear first, while still maintaining the ability to explore less obvious but potentially valuable connections.
Through this architecture, the system achieves both high precision in finding relevant studies and the flexibility to handle the complex relationships inherent in clinical research data. The tight integration between graph structure and vector search capabilities provides a powerful foundation for intelligent search in the medical domain.
Benefits of accelerating iteration speed
The primary objective for clinical research teams is to expedite the identification of successful therapeutic patterns, systematically iterating through various studies to rapidly recognize unsuccessful clinical trials. This helps ensure optimal allocation of time, financial, and material resources throughout the clinical trials and development pipeline.
With an intelligent semantic search system, these teams move from transforming massive amounts of data into a functional knowledge graph and exposing familiar interfaces for various consumers to allow for rapid iteration capability and AI-powered curation to identify non-obvious relationships between data points. Hypermode provides the tightly integrated kits of tools and infrastructure that gives teams building these intelligence search systems, driving outcomes including:
- Operational efficiency. Streamlining database management eliminates the need for separate vector databases such as Pinecone. This consolidation reduces data synchronization complexity while making deployment and maintenance more straightforward. The system also enables real-time updates to both structured and vector data, creating a more responsive and manageable infrastructure.
- Enhanced search capabilities. Dgraph’s capabilities provide a sophisticated approach to data retrieval by combining semantic similarity with structured filtering mechanisms. Its architecture supports multi-vector search with customizable weights, while maintaining context through graph relationships. Users can leverage flexible query generation through LLMs, allowing for more nuanced and precise search results.
- Developer experience. Hypermode’s code-first approach offers a familiar, streamlined experience for developers. Teams benefit from native Go and AssemblyScript support, along with automatic GraphQL API generation. The system seamlessly integrates with existing Python microservices, making it particularly accessible for teams working with diverse technology stacks.
- Scalability and future-proofing. Hypermode's capabilities are built on a foundation of the proven Dgraph infrastructure. Organizations can adopt new features incrementally according to their needs, while maintaining the ability to version ontologies effectively. The platform's architecture facilitates eventual migration to more sophisticated search algorithms, ensuring long-term viability and adaptability.
Looking ahead
The combination of Dgraph's graph database capabilities with the Modus framework provides a robust foundation for building intelligent search systems. This architecture for intelligence search demonstrates how modern graph databases and development frameworks can work together to solve complex search problems while maintaining simplicity in deployment and operations.
The real power of this approach lies in its ability to combine structured graph data with semantic search capabilities, all while keeping the operational complexity manageable. As more organizations face similar challenges in managing and searching complex datasets, this pattern of integrated vector and graph capabilities will become increasingly valuable.
Create powerful, scalable solutions by leveraging Dgraph for querying interconnected datasets and Modus for seamless AI integration. Explore our recipes and adapt these templates for your project!